Submit your event on Induced Pluripotent Stem Cells (iPSCs) to be featured.
Induced Pluripotent Stem Cells (iPSCs)
The discovery of induced pluripotent stem cells (iPSCs), by Takahashi and Yamanaka in 2006, revolutionized the field of stem cell research. iPSCs can provide an unlimited supply of undifferentiated cells from readily available somatic, or differentiated, cells. Prior to this discovery, the primary source of pluripotent stem cells were embryonic stem cells (ESCs), derived from the blastocyst of an existing embryo. iPSCs have paved the way for an expansion in research and clinical areas of regenerative medicine, cell and gene therapy, and disease pathology. What is an iPSC?In their initial experiments, Takahashi and Yamanaka isolated mouse fibroblasts and reprogrammed them into cartilage, neural tissue, and columnar epithelium. More recently, iPSCs have been generated from other somatic cells, including peripheral blood mononuclear cells (PBMCs), keratinocytes, and hematopoietic stem cells (HSCs). Less controversial, and equally pluripotent, somatic cells can be reprogrammed by introduction of four transcription factors Oct4, Sox2, Klf4 and c-Myc, often referred to as “Yamanaka Factors.” The addition of two transcription factors, LIN-28 and Nanog, has been shown to increase reprogramming efficiency. After reprogramming to a “de-differentiated” state, iPSCs can generate virtually any cell type. 
What is an iPSC? Somatic cells can be cultured and reprogrammed into iPSCs using canonical pluripotency transcription factors. iPSCs are differentiated into various tissues for basic research and translational studies. *Though c-Myc is an essential transcription factor for iPSC reprogramming, high oncogenic potential makes it a poor factor to integrate for translational applications. iPSC ReprogrammingThere are four general groups to categorize an iPSC reprogramming strategy: 
Integrative Reprogramming techniques require the reprogramming factors to be inserted permanently into the host cell genome. This strategy can be further divided into viral and non-viral methods. The pioneering iPSC experiments by Yamanaka et al., were conducted by integrating four transgenes using retroviral vectors. ViralThough they are the most efficient method for reprogramming iPSCs, lenti- and retroviral based transduction methods have distinct drawbacks for clinical and translational applications:
Some of these challenges can be addressed with newer technologies. For example, instead of using four separate transgenes with four separate integration sites and, therefore, four chances for mutation, researchers have developed polycistronic lentiviruses, which express all four transgenes sequentially with 2A Peptide self-cleavage sites. Additionally, researchers can flank their transgene insert with loxP sites and use transient transfection with Cre-recombinase to excise the integrated transgenes after reprogramming is complete. 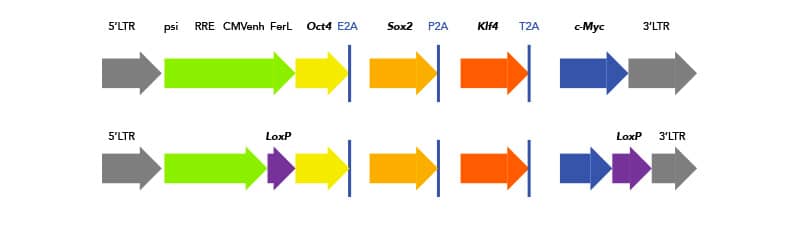
Schematic of polycistronic insert with Yamanaka factors for iPSC reprogramming. (Top) 2A peptide cleavage sites are highlighted. (Bottom) One solution for removing integrated reprogramming genes is flanking loxP sites. Schematic of loxP sites flanking polycistronic insert. Non-viralTransposon technology is a next-generation solution to avoid traditional downfalls of virus transductions. Transposable elements, or transposons, are mobile genomic elements that can move around the genome. In the case of genetic editing and iPSC generation, it refers to a plasmid carrying a transposon with all the reprogramming elements. TcBusterTM is a transposon-based technology developed by Bio-Techne. In addition to reducing mutagenic potential and cargo-size constraints conferred by traditional viral transduction, gene editing with TcBuster is faster and safer for clinical and translational samples. TcBuster technology can also be used to engineer additional genetic changes in iPSCs. Non-integrative Reprogramming techniques are the preferred methodology for clinical and translational iPSC generation. They require no genomic integration, and therefore have significantly reduced chance of introducing harmful mutations. However, non-integrative methods are less efficient, and expression of reprogramming factors is more transient. If non-integrative methods are chosen, there are many small molecules available to increase reprogramming success.
iPSC CharacterizationThe fastest and simplest way to verify successful generation of iPSCs is by morphology, or cell shape. iPSCs should form a culture of tightly packed cells with large nucleus, large nucleoli, and very little cytoplasm. More empirical verification can be done by staining for markers associated with stem cell signaling. After reprogramming, iPSCs should express endogenous pluripotency factors, similar to ESCs. These include both transcription factors, such as Oct4, Sox2, and Nanog, as well as surface markers, like SSEA-4 and TRA-1-60. Interestingly, SSEA-1 is expressed on the surface of mouse, but not human, iPSCs. To maintain and expand iPSCs, cells are traditionally cultured and passaged with feeder cells, such as fibroblasts. For clinical and translational applications, Bio-Techne’s ExCellerate iPSC Expansion Medium is animal-free and removes the need for feeder cells and supplemental growth factors.
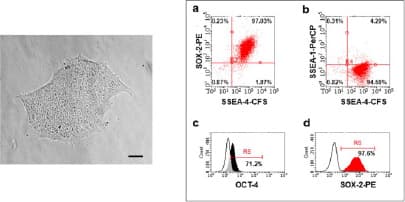
Verification of pluripotency after reprogramming by surface and intracellular flow cytometry. (Left) Morphology- Human iPSC colonies (passage 4) with smooth edges and a non-differentiated phenotype. Magnification: 10x. Scale bar: 80 µm. (Right) Using H/M Pluripotent Stem Cell Multi-Color Flow Cytometry Kit to verify pluripotency of human iPSC multi-color flow cytometry. The majority of cells were positive for (a,d) SOX-2, (a, b) SSEA-4 and (c) OCT-3/4 and were negative for (b) SSEA-1. Image adapted from Harbuzariu, A., Pitts, S., Cespedes, J.C. et al. Modelling heme-mediated brain injury associated with cerebral malaria in human brain cortical organoids. Sci Rep 9, 19162 (2019). https://doi.org/10.1038/s41598-019-55631-8, provided by CC-BY license. 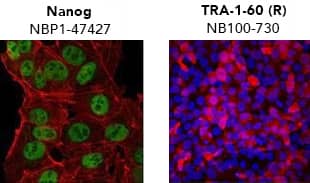
Verification of pluripotency by Immunocytochemistry/Immunofluorescence. (Left) Confocal immunofluorescence analysis of Mouse Anti-Human Nanog Antibody (1E6C4) (Catalog # NBP1-47427) (green). Actin filaments have been labeled with DY-554 phalloidin (red). Nanog staining was confined to the nucleus. (Right) ADLF1 induced pluripotent stem cell line stained with Mouse Anti-Human TRA-1-60 (TRA-1-60) (Catalog # NB100-730) and Anti-Mouse IgG Secondary Antibody (red) and counterstained with DAPI (blue). TRA-1-60 staining was confined to the cell surface. iPSC DifferentiationOne of the most convincing methods to confirm successful reprogramming is to functionally verify pluripotency. To be considered “functionally pluripotent”, iPSCs must be able to differentiate into endoderm, ectoderm, and mesoderm layers. All cell and tissue types develop from one of these layers. Bio-Techne supplies kits to reliably and reproducibly differentiate pluripotent stem cells into cardiomyocytes, neural progenitors, and hepatocytes, in addition to individual growth factors and small molecules. 
Functional Verification of iPSC Pluripotency. iPS2 human induced pluripotent stem cells were differentiated to ectoderm, mesoderm, and endoderm using the Human Pluripotent Stem Cell Functional Identification Kit (Catalog # SC027). The kit also contains antibodies targeting Otx2 (ectoderm), Brachyury (mesoderm), and SOX17 (endoderm) for the confirmation of differentiation status. To further evaluate lineage commitment, cells were stained with a Goat Anti-Human SOX1 Antigen Affinity-purified Antibody (Catalog # AF3369), a Goat Anti-Human HAND1 Antigen Affinity-purified Antibody (Catalog # AF3168), and a Goat Anti-Human HNF-3 beta/FoxA2 Antigen Affinity-purified Antibody (Catalog # AF2400). The cells were stained using the NorthernLights™557-conjugated Donkey Anti-Goat IgG Secondary Antibody (Catalog # NL001; red) and the nuclei were counterstained with DAPI (blue). Explore Solutions for iPSC Reprogramming, Differentiation, and Characterization Applications of iPSCsDisease research often relies on the use of animal models or two-dimensional (2D) in vitro culture systems. Though extremely useful, animal models are limited in their ability to recapitulate complex diseases and accurately model human cellular responses to new drugs and therapies. Traditional in vitro culture systems rely on examining cellular responses in a contrived 2D environment, with cells grown either in a monolayer plastic dish or in suspension surrounded by culture media. Advancements in cell culture techniques to include organoid and 3D cultures that more closely recapitulate in vivo tissue microenvironment, exponentially expand the applications for iPSCs. 
Diverse applications of iPSCs. Somatic cells are harvested from patient and reprogrammed into iPSCs. The resulting patient specific iPSCs can then be used in disease modeling and drug screening to generate disease and patient-specific therapies. Additionally, patient-specific iPSCs can be modified to repair genetic mutations. These repaired iPSCs can then be transplanted into the patient to restore tissue functionality. Select applications: Drug toxicity screening: new therapies and drugs often fail in human trials due to unforeseen toxicity. To limit this, iPSCs can efficiently be differentiated into organoids to screen target drugs for toxicity. Hepatocytes, neurons, and cardiomyocytes are the three most common tissue for drug toxicity screening.
Disease modeling: iPSCs allow for direct studying of disease state from the patient’s own cells. Additionally, researchers can directly alter patient cells via gene editing and determine the impact for the individual patient.
Regenerative Medicine: With the ability to generate disease-free tissues using off-the-shelf or patient-derived cells, iPSCs have a bright future in regenerative medicine.
Limitations of iPSCsDuring somatic cell reprogramming to iPSCs, there are many genetic and epigenetic changes that occur. With current reprogramming technologies, the epigenetic landscape of the somatic cells is often incompletely or aberrantly modified. This phenomenon, known as epigenetic memory, often biases the iPSCs to differentiate toward their cell of origin. New ExCellerate™ iPSC Expansion MediumSupports robust expansion and maintenance of pluripotent stem cell culture for enhanced consistency and reproducibility.
Select ReferencesRaab S, Klingenstein M, Liebau S, Linta L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int 2014;2014:. https://doi.org/10.1155/2014/768391.
Abbar A Al, Ngai SC, Nograles N, Alhaji SY, Abdullah S. Induced Pluripotent Stem Cells: Reprogramming Platforms and Applications in Cell Replacement Therapy. Biores Open Access 2020;9:121. https://doi.org/10.1089/BIORES.2019.0046. Goyak KMO, Laurenzana EM, Omiecinski CJ. Hepatocyte Differentiation. Methods Mol Biol 2010;640:115. https://doi.org/10.1007/978-1-60761-688-7_6. Rajala K, Pekkanen-Mattila M, Aalto-Setälä K. Cardiac Differentiation of Pluripotent Stem Cells. Stem Cells Int 2011;2011:. https://doi.org/10.4061/2011/383709.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||